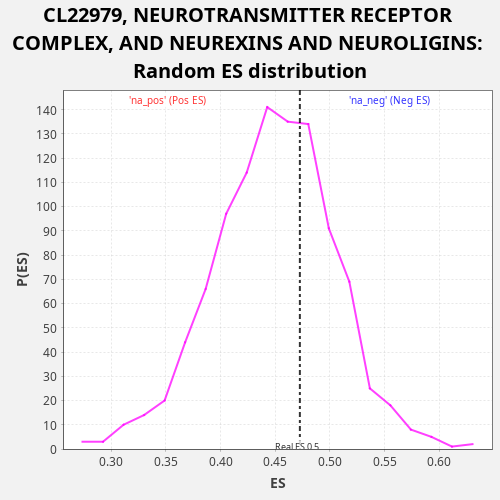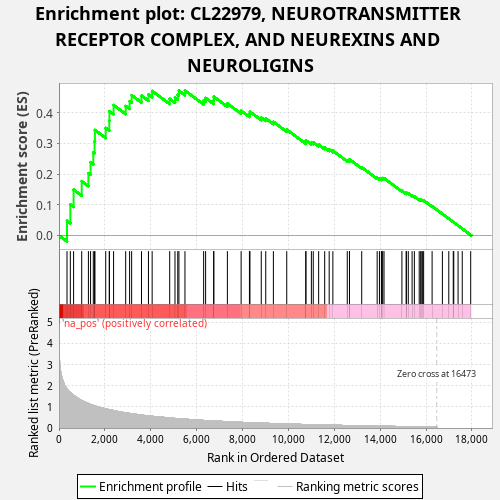
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Irritable bowel syndrome__20002-both_sexes-1154 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL22979, NEUROTRANSMITTER RECEPTOR COMPLEX, AND NEUREXINS AND NEUROLIGINS |
| Enrichment Score (ES) | 0.4724839 |
| Normalized Enrichment Score (NES) | 1.0534552 |
| Nominal p-value | 0.334 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CBLN4 | 349 | 1.841 | 0.0479 | Yes |
| 2 | GRIK1 | 494 | 1.663 | 0.1009 | Yes |
| 3 | NLGN2 | 636 | 1.542 | 0.1495 | Yes |
| 4 | NRXN3 | 990 | 1.291 | 0.1771 | Yes |
| 5 | DLGAP3 | 1282 | 1.153 | 0.2031 | Yes |
| 6 | CBLN2 | 1376 | 1.112 | 0.2386 | Yes |
| 7 | GRIN2C | 1495 | 1.066 | 0.2711 | Yes |
| 8 | GRIN1 | 1551 | 1.048 | 0.3064 | Yes |
| 9 | GRIN2A | 1565 | 1.043 | 0.3439 | Yes |
| 10 | GRIK3 | 2036 | 0.899 | 0.3506 | Yes |
| 11 | HTR3B | 2192 | 0.866 | 0.3737 | Yes |
| 12 | NLGN1 | 2197 | 0.865 | 0.4051 | Yes |
| 13 | GRIA2 | 2379 | 0.823 | 0.4252 | Yes |
| 14 | SHISA7 | 2906 | 0.715 | 0.4220 | Yes |
| 15 | SHANK1 | 3077 | 0.687 | 0.4377 | Yes |
| 16 | GRIN2D | 3166 | 0.673 | 0.4574 | Yes |
| 17 | SHISA6 | 3596 | 0.611 | 0.4558 | Yes |
| 18 | SYNGAP1 | 3899 | 0.568 | 0.4598 | Yes |
| 19 | HTR3C | 4060 | 0.552 | 0.4711 | Yes |
| 20 | GRIA4 | 4825 | 0.470 | 0.4456 | Yes |
| 21 | GRIA1 | 5055 | 0.450 | 0.4493 | Yes |
| 22 | GRM4 | 5176 | 0.440 | 0.4587 | Yes |
| 23 | CNTN4 | 5227 | 0.436 | 0.4719 | Yes |
| 24 | GRIK2 | 5490 | 0.415 | 0.4725 | Yes |
| 25 | FBXO40 | 6304 | 0.356 | 0.4401 | No |
| 26 | OLFM2 | 6390 | 0.351 | 0.4482 | No |
| 27 | CLSTN3 | 6740 | 0.330 | 0.4408 | No |
| 28 | DLG2 | 6749 | 0.329 | 0.4524 | No |
| 29 | HOMER3 | 7339 | 0.296 | 0.4304 | No |
| 30 | NETO1 | 7938 | 0.265 | 0.4067 | No |
| 31 | CDH9 | 8300 | 0.249 | 0.3957 | No |
| 32 | GRM1 | 8323 | 0.248 | 0.4035 | No |
| 33 | CACNG5 | 8816 | 0.228 | 0.3844 | No |
| 34 | NETO2 | 9011 | 0.220 | 0.3816 | No |
| 35 | SLITRK5 | 9344 | 0.208 | 0.3706 | No |
| 36 | MDGA1 | 9926 | 0.188 | 0.3451 | No |
| 37 | CACNG8 | 10756 | 0.160 | 0.3046 | No |
| 38 | HTR3E | 10762 | 0.160 | 0.3102 | No |
| 39 | DLG4 | 11000 | 0.154 | 0.3026 | No |
| 40 | HOMER2 | 11082 | 0.152 | 0.3037 | No |
| 41 | GRM5 | 11317 | 0.146 | 0.2959 | No |
| 42 | GRID1 | 11580 | 0.139 | 0.2864 | No |
| 43 | CHRM5 | 11776 | 0.133 | 0.2804 | No |
| 44 | GRIN3B | 11937 | 0.129 | 0.2762 | No |
| 45 | LRRTM1 | 12564 | 0.114 | 0.2454 | No |
| 46 | HTR3D | 12660 | 0.112 | 0.2442 | No |
| 47 | HTR3A | 12662 | 0.112 | 0.2482 | No |
| 48 | DLGAP4 | 13194 | 0.101 | 0.2222 | No |
| 49 | CNIH3 | 13867 | 0.087 | 0.1879 | No |
| 50 | SHANK3 | 13967 | 0.085 | 0.1855 | No |
| 51 | SHANK2 | 14052 | 0.084 | 0.1839 | No |
| 52 | VWC2L | 14067 | 0.083 | 0.1862 | No |
| 53 | GRIN3A | 14107 | 0.083 | 0.1870 | No |
| 54 | TJAP1 | 14166 | 0.082 | 0.1867 | No |
| 55 | GRM7 | 14946 | 0.068 | 0.1457 | No |
| 56 | NLGN4X | 15135 | 0.065 | 0.1376 | No |
| 57 | NRXN1 | 15139 | 0.065 | 0.1398 | No |
| 58 | NLGN3 | 15224 | 0.064 | 0.1375 | No |
| 59 | HOMER1 | 15390 | 0.060 | 0.1304 | No |
| 60 | NRXN2 | 15489 | 0.058 | 0.1271 | No |
| 61 | CNIH2 | 15700 | 0.054 | 0.1173 | No |
| 62 | DLGAP1 | 15767 | 0.053 | 0.1156 | No |
| 63 | DLG3 | 15784 | 0.052 | 0.1166 | No |
| 64 | SHISA9 | 15849 | 0.051 | 0.1149 | No |
| 65 | PTCHD1 | 15893 | 0.050 | 0.1143 | No |
| 66 | LHFPL4 | 16262 | 0.041 | 0.0952 | No |
| 67 | CBLN1 | 16710 | -0.000 | 0.0703 | No |
| 68 | CHRM3 | 16989 | -0.000 | 0.0547 | No |
| 69 | GRIK4 | 17184 | -0.000 | 0.0439 | No |
| 70 | GRID2 | 17206 | -0.000 | 0.0427 | No |
| 71 | CACNG2 | 17394 | -0.000 | 0.0323 | No |
| 72 | GRM8 | 17576 | -0.000 | 0.0222 | No |
| 73 | GRIN2B | 17944 | -0.000 | 0.0017 | No |